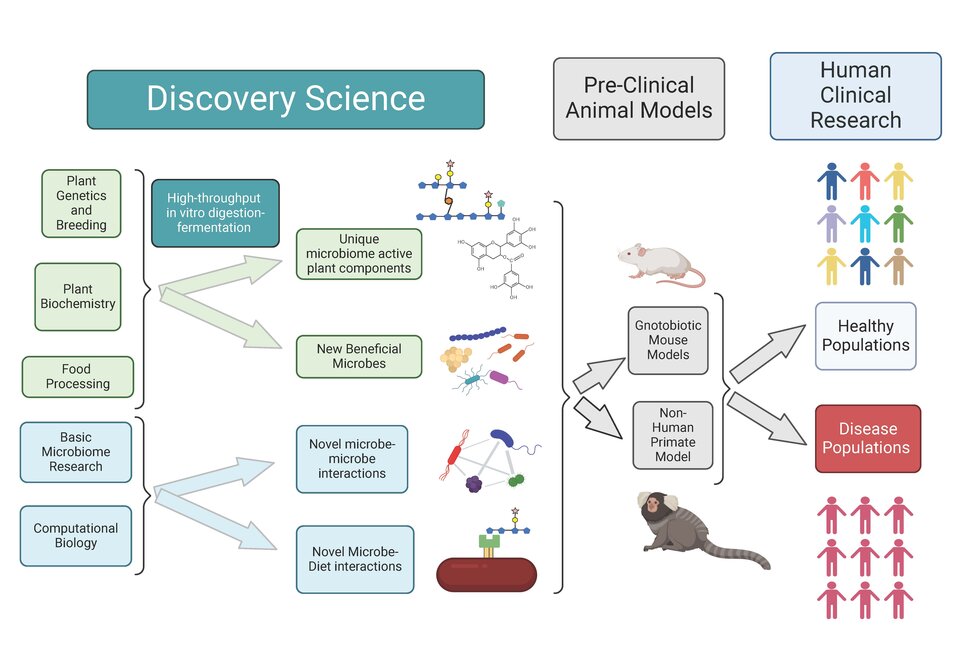
The NFHC research platform unites its member faculty from a diverse array of disciplines to work as collaborative teams and it provides a compass to drive discoveries and fundamental science in the agricultural and food sciences toward pre-clinical and clinical translation. Teams working in discovery science include plant geneticists, biochemists, microbiologists, food scientists, and computational biologists and they collectively drive discovery products such as
- novel components in food crops that affect the human gut microbiome,
- new combinations of dietary components that stimulate growth of beneficial gut microbes
- unique interactions among microbial species in the microbiome that are targets for dietary manipulation (e.g. consortia of primary degraders and secondary feeders)
- unique pathways through which individual microbes degrade specific food components.
- How food processing methods such as cooking, extrusion, and fermentation affects the bioavailability of carbohydrates for gut microbiome fermentation
NFHC microbiologists and immunologists study fundamental aspects of the microbiome, including structural biology of individual gut microbes, ecological characteristics of the gut microbiome as well as interactions of enteric pathogens such as C. dificile with the gut microbiome. Examples include:
- The roles of extracellular structures such as flagella and type IV pili biofilm formation by C. dificile
- The roles of antibiotics, bile salt metabolism, and amino acid metabolism on colonization by C. dificile
- Niche priority and niche occupation by gut bacterial species, and fitness advantages of native gut bacteria
The NFHC platform also includes bioinformatics and computational biology, where faculty collaborate to mine complex sets of genomic, metagenomic, microbiome, and metabolome data to study the types of substrates that might be produced by a given plant as well as how specific substrates can be degraded by individual gut microbes or consortia of gut microbes. Examples of major research outcomes relating to bioinformatics and computational biology include:
- Development and support for the dbCAN3 database/server for predicting substrates for CAZyme gene clusters in microbial genomes and their substrates
- Genome sequencing of wild cacoa
- Structure and linkages of complex substrates drives substrate utilization and microbial community structure
[Right click image and open in new tab to view larger.]
Research products emerging from the discovery teams are then evaluated for their translational potential by collaborative teams of microbiologists, immunologists, food scientists, nutritionists, and primatologists in pre-clinical animal models. For example, collaborative teams use:
- feeding studies of different genotypes of a food crop to confirm effects on the microbiome of gnotobiotic mice or common marmosets are similar as those from in vitro studies used in discovery
- feeding studies to demonstrate how different genotypes of a food crop influence disease outcomes such as colitis in the gnotobiotic mouse model
- effects of a candidate microbial species on susceptibility to disease such as colitis in a gnotobiotic mouse model
- feeding studies to determine if a specific dietary component can influence behavior outcomes in the marmoset model through its effects on the gut microbiome
Candidate foods and food ingredients with desirable microbiome activity are finally evaluated for translation and efficacy in human clinical trials. Here, clinicians such as gastroenterologists work with nutritionists, microbiologists, and immunologists to evaluate how a microbiome-active food affects the human gut microbiome and outcomes related to health such as blood biomarkers of inflammation or metabolic markers such as blood sugar or triglycerides. Studies can be done in different populations, including healthy persons or individuals with complex diseases such as inflammatory bowel disease or metabolic diseases that are known to be associated with dysbiosis in the gut microbiome.